TRACTION STRESS ANALYSIS AND MODELING REVEAL AMOEBOID MIGRATION IN CONFINED SPACES IS ACCOMPANIED BY EXPANSIVE FORCES AND REQUIRES THE STRUCTURAL INTEGRITY OF THE MEMBRANE-CORTEX INTERACTIONS
by Ai Kia Yip, Keng-Hwee Chiam and Paul Matsudaira
Integrative Biology, 2015, DOI: 10.1039/C4IB00245H. First published online 27 May 2015
Cell migration is crucial in many biological processes such as embryonic development, wound closure, as part of the body’s inflammatory response, as well as during cancer cell metastasis. A cell can translocate across a surface by either a mesenchymal or amoeboid cell migration mechanism. While cells are known to adhere to the extracellular matrix and generate contractile cell traction forces during integrin-dependent mesenchymal migration, little is known about the nature of the traction forces required for amoeboid migration in the absence of cell-matrix adhesions.
In this paper, we have combined 3-dimensional traction force microscopy with a confinement assay, where neutrophil-like cells are confined between two pieces of polyacrylamide gels. In the absence of cell-matrix adhesions, confined cells migrate by amoeboid “chimneying”, that is, generate traction by applying expansive and divergent forces against opposing surfaces. This chimneying speed was shown, both experimentally and computationally, to depend on both the magnitude of the intracellular pressure and the location where blebs are formed as determined by the membrane-cortex adhesion strength.
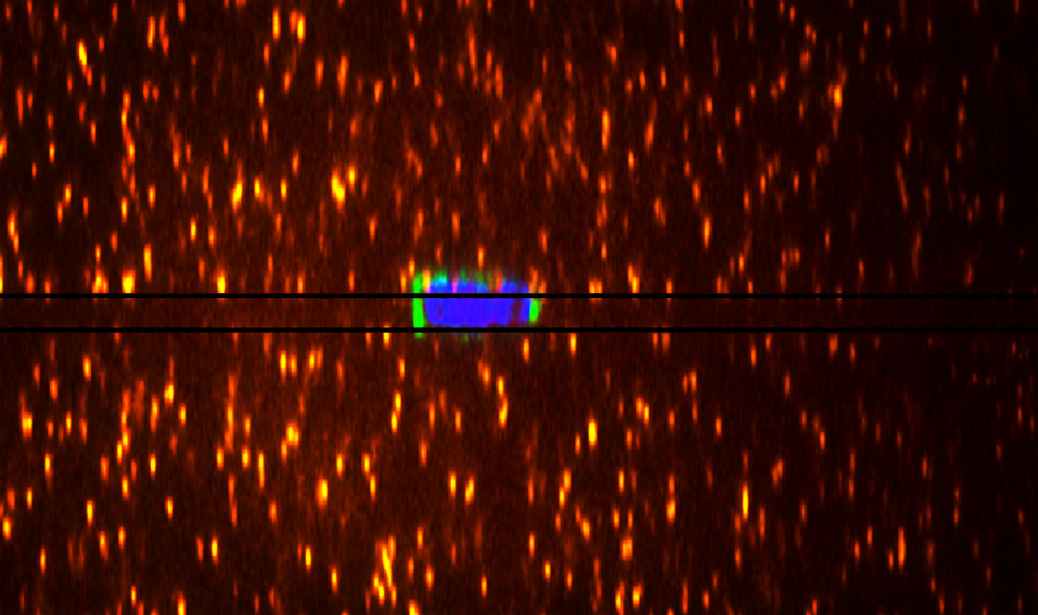
Picture caption: A cell confined between two pieces of non-adhesive polyacrylamide gels. 3D traction forces can be calculated based on gel deformations detected by the displacements of the embedded beads (orange).
Movie caption: An amoeboid neutrophil-like, differentiated HL60 cell migrating in between two pieces of non-adhesive polyacrylamide gels (DIC image, far left). Cell nuclei were stained with Hoechst (middle left panel, blue) and cells were transfected with Lifeact-GFP (middle right panel, green), which labels the F-actin. The three channels are overlaid in the far right panel. The number on the top of the images indicates the time in hour: minute: second.
Learn more about Paul Matsudaira’s research.