
PHUA SIEW CHENG
Assistant Professor
Department of Biological Sciences
National University of Singapore
16 Science Drive 4
Singapore 117558
Singapore
Lab webpage: Neuron Signaling Lab
Academic Qualifications & Training
2021-now Assistant Professor, National University of Singapore
2021-2021 Senior Research Fellow, Institute of Molecular and Cell Biology
2018-2021 Research Fellow, Singapore Bioimaging Consortium
2011-2017 Doctor of Philosophy, John Hopkins University School of Medicine
2010-2011 Research Officer, Institute of Medical Biology
2006-2010 Bachelor of Science (1st Class Honors), Nanyang Technological University
Research Areas
Molecular & systems neuroscience, behavior states, in vitro & in vivo imaging, molecular tools, proteomics.
Research Interests

Neuron signaling in behavior: In my lab, we are fundamentally curious about how our brains form & switch between behavioral states.
The term “behavioral states” could refer to circadian behavior rhythms such as sleep-wake transition, emotional states such as fear & anxiety, & pathological states such as Parkinsonism. Building on behavioral paradigms established by us & others, we employ advanced in vivo imaging, optogenetic & genome editing approaches to illuminate the molecular signaling events in neurons that shape & modulate behavioral states. These approaches are supplemented by molecular tool development, in vitro imaging, proteomics & whole-brain light-sheet microscopy. Our current focus is to unravel the molecular pathways in our two favorite organelles [the neuronal nucleus & the neuronal cilium] that instruct gene expression patterns ultimately encoding brain behavioral states. Through our cross-scale approaches, we aim to bridge the knowledge gap between molecular & systems neuroscience & form a better understanding of how behavior may become awry. 
The neuronal nucleus: Harboring the genome, the nucleus of each neuron is the master console of neuronal activity that ultimately form and shape brain behavioral states. Despite vast knowledge of signaling pathways that modulate gene expression, the dynamics of nuclear signaling during behavior has not been under the spotlight. We are combining advanced in vivo imaging approaches with molecular tool development to visualize and manipulate signaling events in the neuronal nucleus.
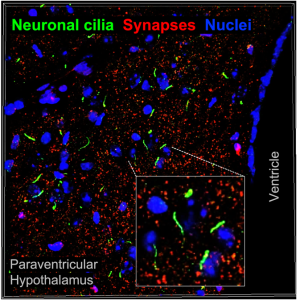
The neuronal cilium: Proposed as the antenna of a cell, the primary cilium of neurons (& thus neuronal cilium) is a ubiquitous solitary hair-like organelle that is under-appreciated in neurobiology. Recent studies have discovered diverse molecular receptor enrichment (e.g. receptors for serotonin, dopamine, melanin-concentrating hormone) in the neuronal cilia but the significance of such ciliary enrichment has not been elucidated. We are applying proteomics and genome editing approaches to understand how primary cilia modulate the activity state of neurons and neural circuits. Moreover, we are intrigued by the commonalities between the cilium and nucleus and are developing molecular approaches to ‘listen’ to the molecular conversations between them.
Awards
2021-2024 National Medical Research Council Young Individual Research Grant
2017 Weintraub Graduate Student Award (Fred-Hutch Cancer Research Center)
2016 The Hans Prochaska Research Award (Johns Hopkins Medicine)
2011-2016 A*STAR National Science Scholarship
2008-2010 A*STAR Pre-Graduate Scholarship
2007 NTU President Research Scholarship
Selected Publications
For full list of publications, please refer to Google Scholar.
Phua, S.C.#, Tan, Y.L.#, Kok, A.M.Y., Senol, E., Chiam, C.J.H., Lee, C.Y., Peng, Y., Lim, A.T.J., Mohammad, H., Lim, J.X., Fu, Y#. A distinct parabrachial to lateral hypothalamus circuit for motivational suppression of feeding by nociception. Science Advances 2021; 7(19), eabe4323. (#co-correspondence)
Jia, S., Phua, S.C., Nihongaki, Y., Li, Y., Pacella, M., Li, Y., Mohammad, A.M., Sun, S., Inoue, T., Schulman, R. Growth and site-specific organization of micron-scale biomolecular devices on living mammalian cells. Nature Communications 2021; 12(5279).
Phua, S.C.*, Nihongaki, Y.*, & Inoue, T. Autonomy declared by primary cilia through compartmentalization of membrane phosphoinositides. Current Opinion in Cell Biology 2018; 50, 72–78. (*co-first authors)
Phua S.C.#, Chiba S., Suzuki M., Su E., Roberson E.C., Pusapati G.V., Setou M., Rohatgi R., Reiter J.F., Ikegami K.#, Inoue T.# Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 2017; 168(1-2), 264-279. (#co-correspondence)
Phua S.C.#, Lin Y.C., Inoue T#. An intelligent nano-antenna: Primary cilium harnesses TRP channels to decode polymodal stimuli. Cell Calcium 2015; 58(4), 415-422. (#co-corresponding authors)
Garcia-Gonzalo F.R.*, Phua S.C.*, Roberson E.C., Garcia III G., Abedin M., Schurmans S., Inoue T., Reiter J.F. Ciliary phosphoinositides modulate Hedgehog signaling. Developmental Cell 2015; 34(4), 400-409. (*co-first)
Su S.*, Phua S.C.*, DeRose R., Chiba S., Narita K., Kalugin P.N., Katada T., Kontani K., Takeda S., and Inoue T. Genetically encoded calcium indicator illuminates calcium dynamics in primary cilia. Nature Methods 2013; 10(11), 1105-1107. (*co-first)
Phua S.C., Pohlmeyer C., Inoue T. Rapidly relocating molecules between organelles to manipulate small GTPase activity. ACS Chemical Biology 2012; 7(12): 1950-1955.
