Dynamics of multiprotein complexes in cellular signalling
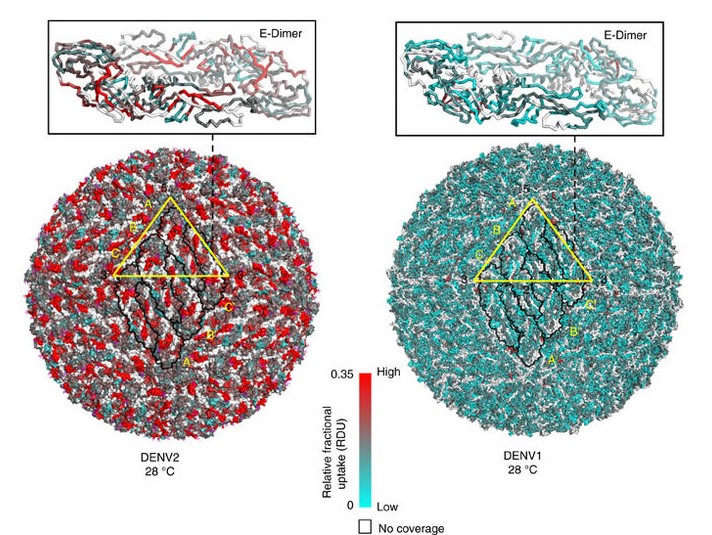
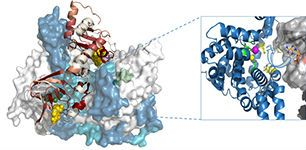
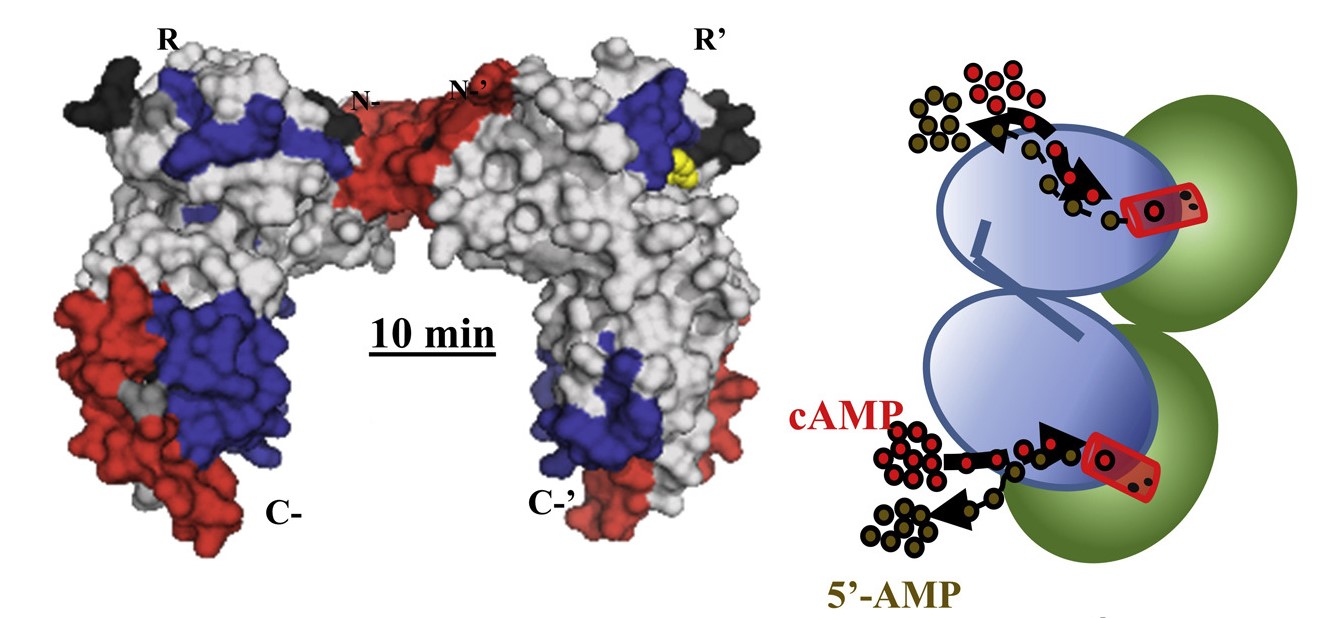
Our research focuses on describing the effects of reversible, covalent phosphorylation (His-Asp two component signalling) and noncovalent phosphates (cyclic nucleotide monophosphate (cAMP), ATP, ADP and AMP) on protein conformational dynamics. Ongoing research areas include role of phosphodiesterases (PDEs) in cAMP homeostasis and allosteric effects of histidine phosphorylation and phosphotransfer to aspartates in two-component signalling.