Our research interests
The main area of research in our lab is in the field of Plant Morphogenesis. We focus on molecular developmental biology with emphasis on phytohormone signaling. The gibberellin signaling intermediate, RGL2 and the cytokinin signaling intermediates (histidine phosphotransfer proteins or AHP and response regulators or ARR) are being studied. The overall aim of our studies is to understand how they control vegetative shoot development. We use rice and Arabidopsis as our main experimental systems for these studies. Another area of research work involves attempts to understand the salt secretion mechanism in the leaves of the mangrove tree species Avicennia.
Recent accomplishments in understanding the regulation of plant morphogenesis
- Work on phytohormone signaling: Our current work is aimed at understanding how plant hormone signaling intermediates regulate plant development. The research focus is on the gibberellin signaling intermediate RGL2 and Arabidopsis histidine phosphotransfer protein (AHP) and Arabidopsis Response Regulator (ARR). We use rice and Arabidopsis as the main experimental systems for these studies.
- In the past we had identified a gene encoding for a cytokinin binding protein (originally isolated from petunia) that is involved in regulating branching, flowering time and adventitious shoot induction from leaf explants. This can be an important candidate gene for developmental enhancement of leafy vegetables where profuse branching and delayed flowering phenotype are highly desirable. This can also contribute to plant biotechnology in the long-term.
- Arising from the above research project and also from our earlier work on ethylene, we have initiated some work on attempts to isolate phytohormone signal transduction intermediates. Recent literature indicates that ethylene and cytokinin signal transduction proceeds by the two-component histidine kinases. Several downstream intermediates have also been isolated by other workers. These studies can contribute towards a basic understanding of phytohormone signal transduction during plant development.
Studies such as these help to identify genes for future crop improvement efforts by genetic enhancement of development either by marker assisted plant breeding or by genetic engineering.
- Work on MADS box genes: We isolated and characterized a floral MADS-box gene, namely, RcMADS1 from Rafflesia cantleyi. RcMADS1 shares sequence similarity with AGAMOUS-LIKE 24 (AGL24) and SHORT VEGETATIVE PHASE (SVP) of Arabidopsis. Ectopic expression of RcMADS1 in Arabidopsis caused early flowering and conversion of sepals and petals into leaf-like structures, and carpels into inflorescences. Using heterologous expression analysis in Arabidopsis, we showed that RcMADS1 is a functional ortholog of Arabidopsis AGL24 in the holoparasitic plant Rafflesia cantleyi Solms-Laubach (Rafflesiaceae).(Ramamoorthy et al. (2013) PLoS ONE 8(6): e67243. doi:10.1371/journal.pone.0067243).
- Previously, we had isolated and characterized some genes that are associated with adventitious shoot regeneration. These include a MADS-box cDNA (PkMADS1) from Paulownia kawakamii, which regulates vegetative shoot development and in vitro shoot regeneration from leaf explants (Plant Journal 2002, 29:141-151). Functional analyses based on transgenic plant strategy revealed that PkMADS1 is a novel MADS box protein that regulates vegetative shoot development in Paulownia. Most of the earlier information on MADS box genes was on regulation of flower development. Our work was one of the first to establish a regulatory role for the vegetatively expressed MADS box proteins in plant development.
- Other work on MADS box genes we have completed include the discovery of AGAMOUS-LIKE24 (AGL24) in regulating flowering time in Arabidopsis (PNAS, USA 2002, 99: 16336-16341). Our work established that AGL24 acts as a key intermediary for integrating the flowering signal between LEAFY and SOC1.
- Another MADS box gene we have cloned and characterized is from the primitive gymnosperm Cycas – which has established that the AGAMOUS gene evolved at least 300 million years ago and that its structure and function have been conserved for that period in all the seed plants (Plant Journal 2004, 37: 566-577).
Mechanism of salt secretion in a mangrove tree species, Avicennia: We are working on various aspects of salt secretion in the leaves of Avicennia, a mangrove tree species. This is approached by physiological, molecular investigations. Differential transcriptomics and proteomics are also attempted in order to identify the molecular regulation of salt secretion.
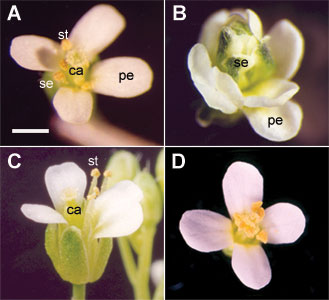 |
Phenotypes of expression of the AGAMOUS ortholog CyAG from Cycas (a gymnosperm) in Arabidopsis (Plant J. 2004, 37:566-577) (A) Wild-type Arabidopsis flower. (B) ag-2 mutant flower. (C) Flowers of ag-2 harboring AGenhancer::D35S::CyAG showing that the stamens and carpel are rescued (four stamens). (D) Flowers of ag-2 harboring AGenhancer::D35S::CyAG showing that the mutant is fully rescued (six stamens). se, sepal; pe, petal; st, stamen; ca, carpel. Scale bar = 1 mm. |
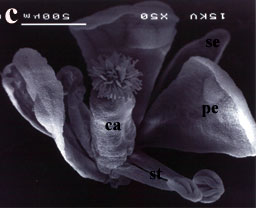 |
SEM image of an Arabidopsis flower harboring CyAG cDNA in the ag-2 mutant background. Note full rescue of the mutant phenotype. |
- Past work on DNA markers: A minor area of research interest in our lab was to develop DNA markers such as AFLP (phylogeny of Johannesteijsmannia - a palm species) and microsatellites (population dynamics in tropical moss species such as Acanthorrhynchium).
 |
We have developed microsatellite markers for a tropical moss, Acanthorrhynchium papillatum |